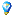https://biomodal.com/blog/how-dna-methylation-affects-gene-expression/DNA
methylationWiki serves as a pivotal epigenetic mechanism that significantly influences gene regulation by recruiting proteins that facilitate gene repression or by hindering transcription factors from attaching to DNA.
This process, alongside histone modifications, forms the core of what is gene expression and plays a fundamental role in molecular biology and genetics, impacting development, differentiation, and the onset of various diseases.
Understanding DNA methylation and its effects on gene expression is vital for advancing our knowledge in the fields of health and disease.
It not only sheds light on the underlying mechanisms of gene regulation but also opens avenues for research into therapeutic interventions for genetic and epigenetic disorders.
● Mechanisms of DNA methylation
DNA methylation is a sophisticated epigenetic mechanism crucial for regulating gene expression in eukaryotic organisms.
This section delves into the intricate processes and enzymes involved in DNA methylation, highlighting its significance in gene regulation.
● DNA methylation process
1. Transfer of methyl groups: the process begins with the transfer of a methyl group to the 5th carbon in the cytosine ring in DNA, forming 5-methylcytosine. This transfer is facilitated by DNA methyltransferases (DNMTs).
2. Role of DNA methyltransferases: DNMTs such as DNMT1, DNMT3a, and DNMT3b play pivotal roles. DNMT1 is responsible for maintaining methylation patterns, while DNMT3a and DNMT3b add methyl groups to DNA during early development.
3. CpG sites targeting: methylation predominantly occurs at CpG sites, though non-CpG methylation exists and is significant in embryonic stem cells.
● Patterns and detection
Erasure and re-establishment: methylation patterns are not permanent; they are largely erased and re-established between generations, ensuring genetic diversity and proper development.
Detection techniques: various techniques such as methylation-specific PCR (MSP) and whole genome bisulfite sequencing (BS-Seq) are employed to detect DNA methylation patterns, providing insights into the methylation status across the genome.
● Functional implications of DNA methylation
Gene expression regulation: by modifying the cytosine bases of DNA, methylation alters the genetic expression. Areas rich in CpG dinucleotides, particularly promoters and enhancers, are key targets for methylation which can suppress gene activity.
Developmental changes: during different developmental stages, the DNA methylation landscape of the genome is dynamically altered, balancing between methylation and demethylation.
Tissue-specific patterns: each cell type inherits a unique methylation pattern, which is crucial for regulating tissue-specific gene expression.
Epigenetic inheritance and imprinting
Genomic imprinting: methylation plays a central role in genomic imprinting, where only one allele of a gene is expressed while the other is silenced epigenetically through methylation.
This imprinting is crucial for normal development and is meticulously regulated during gamete formation and after fertilisation.
● Interaction with histone modifications
Collaborative epigenetic regulation: DNA methylation works in concert with histone modifications to maintain the epigenetic landscape, ensuring that specific patterns are inherited by progeny cells, thus influencing gene expression across generations.
This detailed exploration of the mechanisms of DNA methylation underscores its critical role in gene regulation and its complex interplay with other epigenetic factors.
The article continues ...